Alex C. Morehead
Hopper Postdoctoral Fellow in Machine Learning & Computing Sciences @ Berkeley Lab
About
Welcome! ![]()
I am a Hopper Postdoctoral Fellow at UC Berkeley’s Lawrence Berkeley National Laboratory. Previously, I received my Ph.D. in Computer Science at the University of Missouri, advised by Dr. Jianlin Cheng within the Bioinformatics & Machine Learning Lab. Prior to my Ph.D., I completed my B.S. in Computer Science at Missouri Western State University. My interests in computer science and machine learning began through many fun experiences building and experimenting with bleeding-edge software early on, and that trend continues to this day.
Research
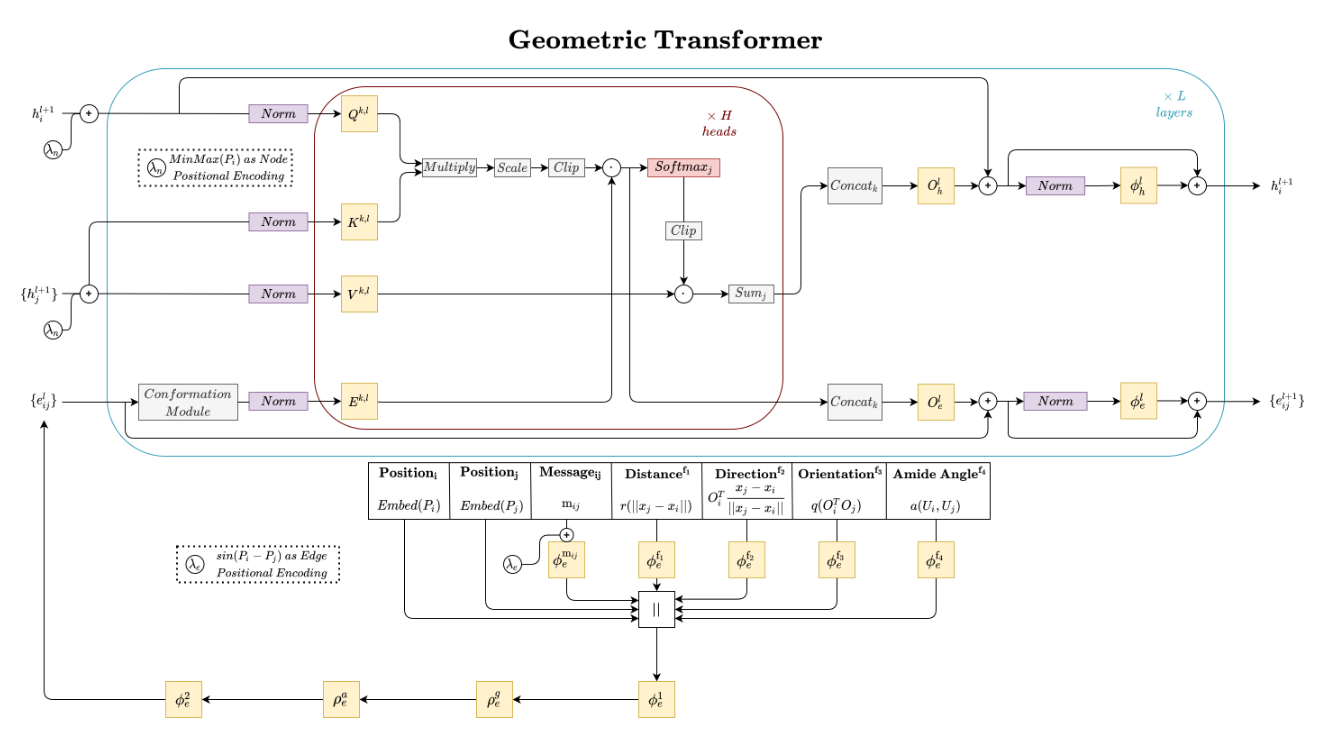
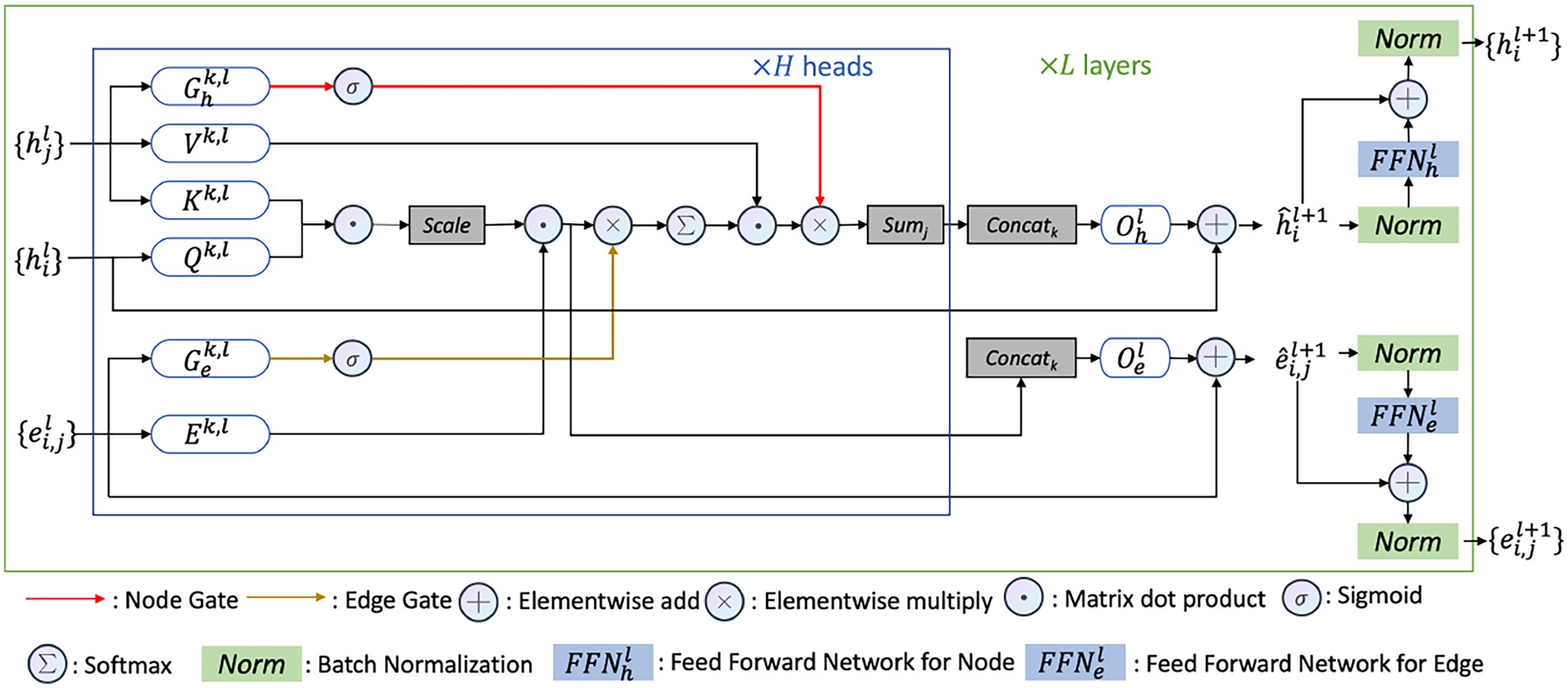
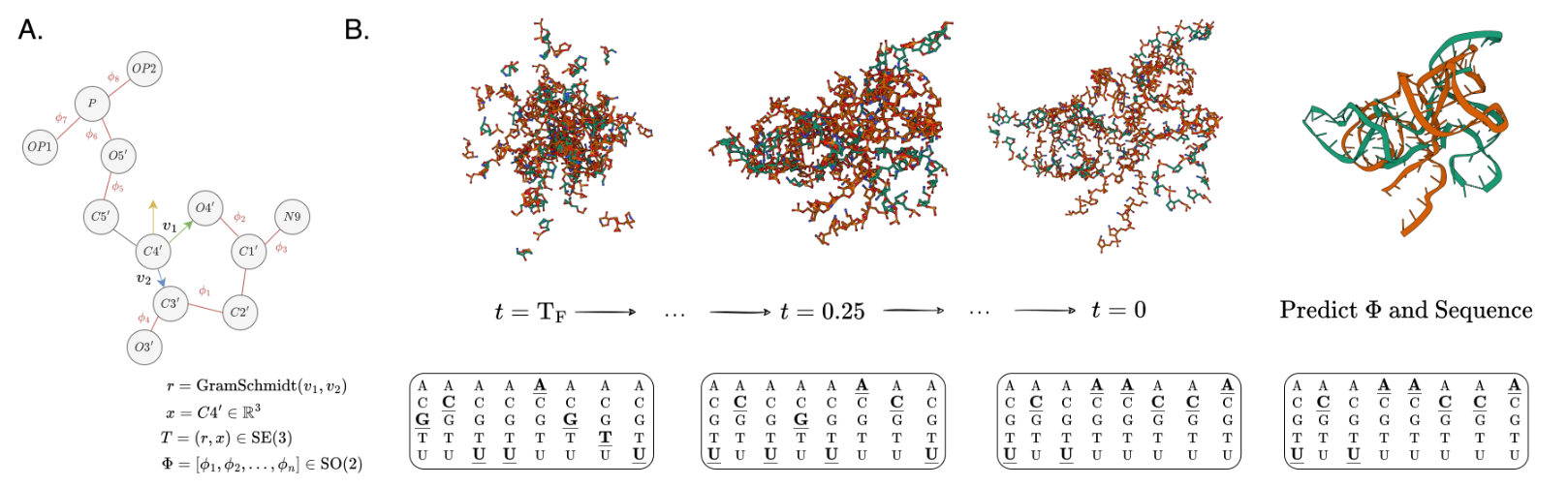
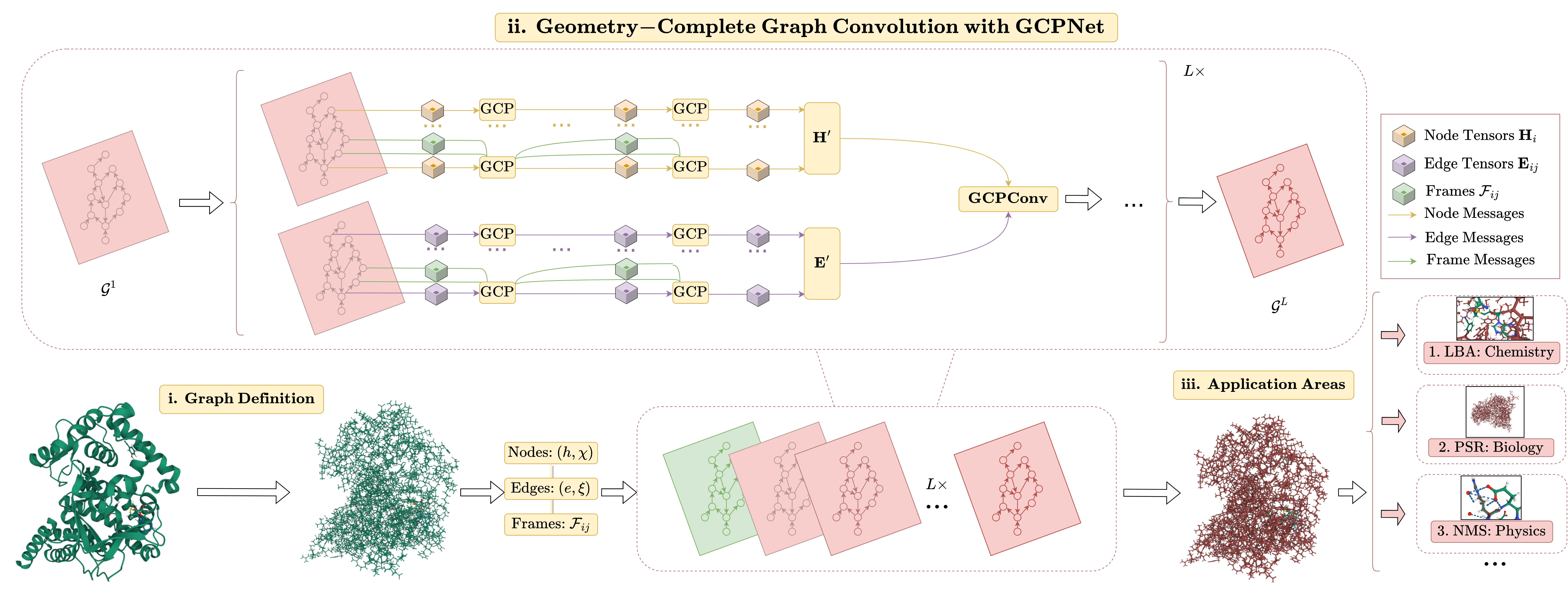
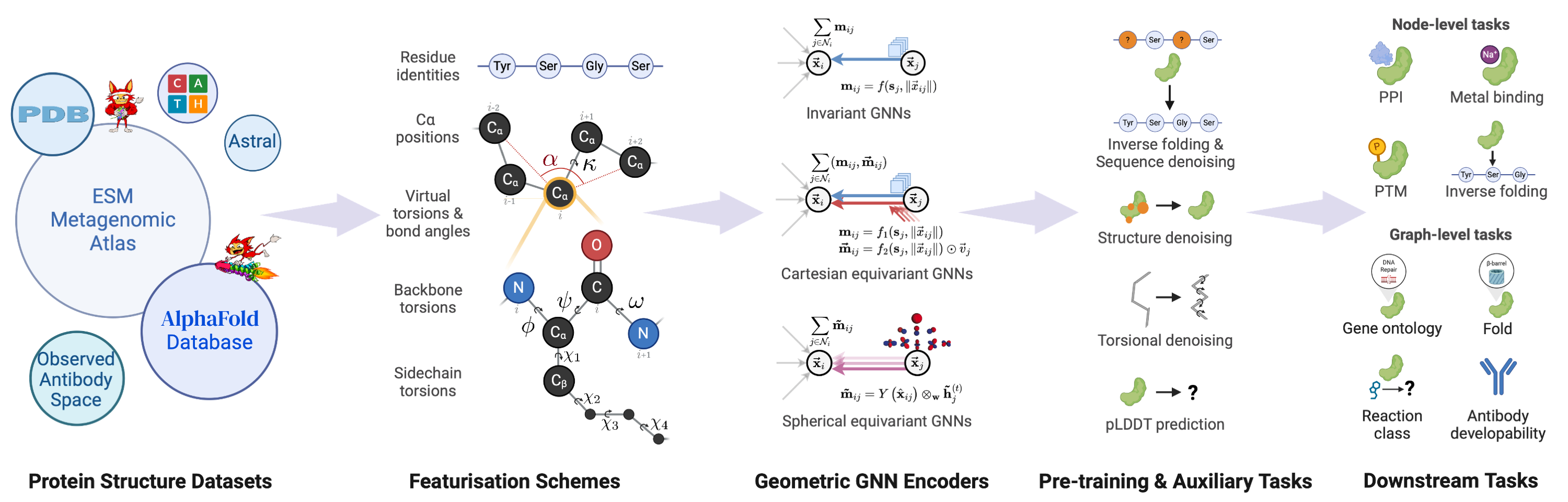
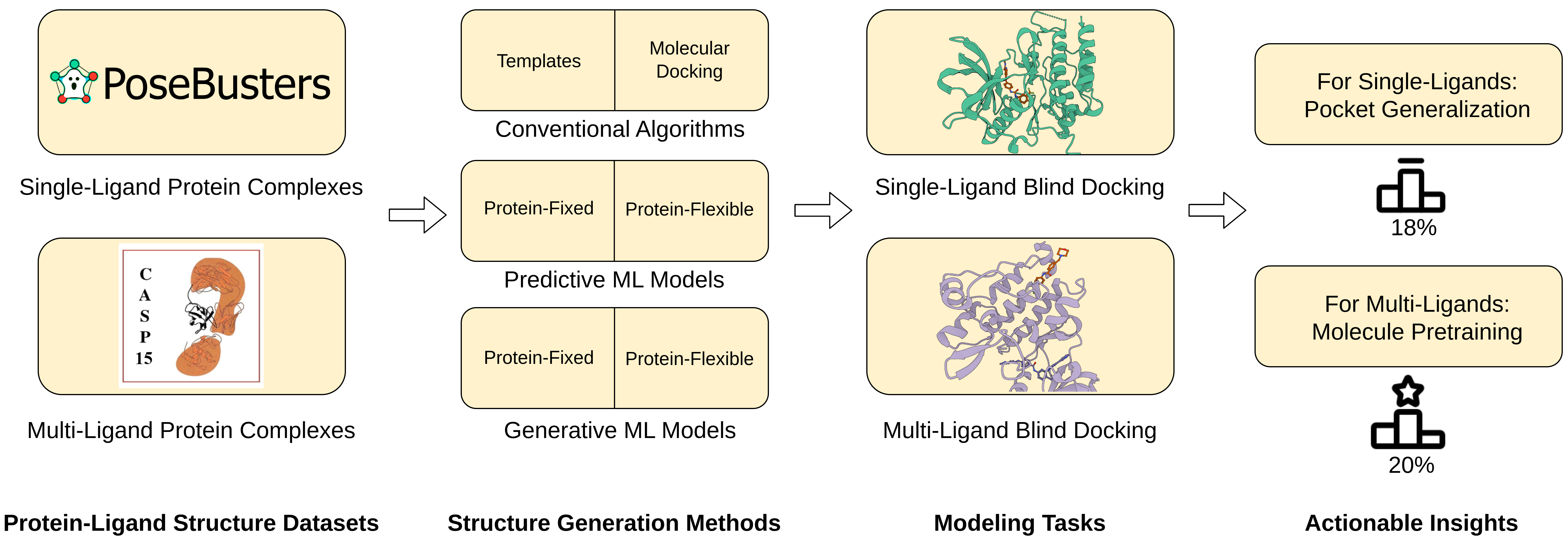
My current research interests include machine learning, deep learning, computational biology, and generative modeling. In particular, I have developed new geometric graph neural network architectures such as GCPNet and the Geometric Transformer for modeling biomolecules of various sizes (e.g., proteins), introduced GCDM for diffusion generative modeling of 3D molecules, assembled PoseBench, the first deep learning benchmark for broadly applicable protein-ligand docking, and released FlowDock, the first all-atom flow matching model for protein-ligand structure and binding affinity prediction.
Writing
I enjoy sharing thoughts on literature, research, and applications on my blog.
Education
Looking to get started with machine learning?
Mentorship/Collaboration
Interested in chatting AI or active research problems?
news
| Oct 23, 2025 | One paper accepted by Nature Machine Intelligence |
|---|---|
| Jul 20, 2025 | New review paper on flow matching models and their applications in bioinformatics |
| Jun 02, 2025 | Honored to start researching at Berkeley Lab as a Hopper Postdoctoral Fellow! |
| Apr 28, 2025 | Successfully defended my PhD dissertation, with a PDF and slides available! |
| Apr 07, 2025 | One paper accepted by ISMB |
| Jan 22, 2025 | One spotlight accepted by ICLR |
| Dec 03, 2024 | Presented MULTICOM_ligand & FlowDock as top-ranked DL methods at CASP16 |
| Jul 03, 2024 | One paper accepted by Nature CommsChem |
| Jun 17, 2024 | Two ICML workshop papers selected for spotlight presentations |
| Feb 19, 2024 | One paper accepted by Bioinformatics |
| Jan 16, 2024 | One paper accepted by ICLR |
| Jul 25, 2023 | Published and presented one paper at ISMB |
| Jan 20, 2022 | One paper accepted by ICLR |
latest posts
| Jan 04, 2026 | Living values |
|---|---|
| Dec 07, 2025 | On metacognition |
| Nov 02, 2025 | The intersection of tools, introspection, and discovery |